Mol:FL63AGNS0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5545 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5545 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5545 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5545 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0160 -0.1583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0160 -0.1583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4774 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4774 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4774 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4774 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0160 1.0854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0160 1.0854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9389 -0.1583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9389 -0.1583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4004 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4004 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4004 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4004 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9389 1.0854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9389 1.0854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0927 1.0852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0927 1.0852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1106 -0.1424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1106 -0.1424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0792 1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0792 1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6239 0.7369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6239 0.7369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1686 1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1686 1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1686 1.6803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1686 1.6803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6239 1.9948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6239 1.9948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0792 1.6803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0792 1.6803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0160 -0.7798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0160 -0.7798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7128 1.9945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7128 1.9945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6239 2.6232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6239 2.6232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7128 0.7372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7128 0.7372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1106 -0.7322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1106 -0.7322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3717 -1.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3717 -1.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6203 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6203 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6203 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6203 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2063 -2.0416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2063 -2.0416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7924 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7924 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7924 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7924 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2063 -0.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2063 -0.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3782 -2.0415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3782 -2.0415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7848 -2.2107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7848 -2.2107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4992 -2.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4992 -2.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3782 -0.6882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3782 -0.6882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0927 -1.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0927 -1.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 1 11 1 0 0 0 0 | + | 1 11 1 0 0 0 0 |
| − | 8 12 1 6 0 0 0 | + | 8 12 1 6 0 0 0 |
| − | 9 13 1 6 0 0 0 | + | 9 13 1 6 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 17 21 1 0 0 0 0 | + | 17 21 1 0 0 0 0 |
| − | 15 22 1 0 0 0 0 | + | 15 22 1 0 0 0 0 |
| − | 12 23 1 0 0 0 0 | + | 12 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 29 34 1 0 0 0 0 | + | 29 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 32 33 | + | M SAL 1 2 32 33 |
| − | M SBL 1 1 35 | + | M SBL 1 1 35 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 35 -7.7363 4.5654 | + | M SBV 1 35 -7.7363 4.5654 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 37 | + | M SBL 2 1 37 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 37 -6.7289 5.0727 | + | M SBV 2 37 -6.7289 5.0727 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL63AGNS0006 | + | ID FL63AGNS0006 |
| − | KNApSAcK_ID C00008884 | + | KNApSAcK_ID C00008884 |
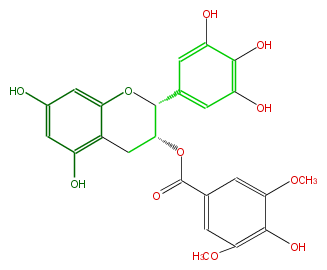
| − | NAME Epigallocatechin 3-O-(3,5-di-O-methylgallate) | + | NAME Epigallocatechin 3-O-(3,5-di-O-methylgallate) |
| − | CAS_RN 173484-92-9 | + | CAS_RN 173484-92-9 |
| − | FORMULA C24H22O11 | + | FORMULA C24H22O11 |
| − | EXACTMASS 486.116211546 | + | EXACTMASS 486.116211546 |
| − | AVERAGEMASS 486.42488000000003 | + | AVERAGEMASS 486.42488000000003 |
| − | SMILES O(C(C3)C(Oc(c4)c3c(cc(O)4)O)c(c2)cc(c(c(O)2)O)O)C(=O)c(c1)cc(OC)c(c(OC)1)O | + | SMILES O(C(C3)C(Oc(c4)c3c(cc(O)4)O)c(c2)cc(c(c(O)2)O)O)C(=O)c(c1)cc(OC)c(c(OC)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-2.5545 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5545 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0160 -0.1583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4774 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4774 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0160 1.0854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9389 -0.1583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4004 0.1527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4004 0.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9389 1.0854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0927 1.0852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1106 -0.1424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0792 1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6239 0.7369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1686 1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1686 1.6803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6239 1.9948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0792 1.6803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0160 -0.7798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7128 1.9945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6239 2.6232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7128 0.7372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1106 -0.7322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3717 -1.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6203 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6203 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2063 -2.0416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7924 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7924 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2063 -0.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3782 -2.0415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7848 -2.2107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4992 -2.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3782 -0.6882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0927 -1.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
1 11 1 0 0 0 0
8 12 1 6 0 0 0
9 13 1 6 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
3 19 1 0 0 0 0
16 20 1 0 0 0 0
17 21 1 0 0 0 0
15 22 1 0 0 0 0
12 23 1 0 0 0 0
23 24 2 0 0 0 0
23 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 30 2 0 0 0 0
30 25 1 0 0 0 0
28 31 1 0 0 0 0
27 32 1 0 0 0 0
32 33 1 0 0 0 0
29 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 32 33
M SBL 1 1 35
M SMT 1 OCH3
M SBV 1 35 -7.7363 4.5654
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 37
M SMT 2 OCH3
M SBV 2 37 -6.7289 5.0727
S SKP 8
ID FL63AGNS0006
KNApSAcK_ID C00008884
NAME Epigallocatechin 3-O-(3,5-di-O-methylgallate)
CAS_RN 173484-92-9
FORMULA C24H22O11
EXACTMASS 486.116211546
AVERAGEMASS 486.42488000000003
SMILES O(C(C3)C(Oc(c4)c3c(cc(O)4)O)c(c2)cc(c(c(O)2)O)O)C(=O)c(c1)cc(OC)c(c(OC)1)O
M END