Mol:FL7AAGGL0069
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.4461 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4461 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4461 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4461 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1606 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1606 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8750 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8750 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8750 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8750 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1606 0.8155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1606 0.8155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5895 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5895 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3040 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3040 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3040 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3040 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5895 0.8155 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 2.5895 0.8155 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0805 0.8514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0805 0.8514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7950 0.4389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7950 0.4389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5095 0.8514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5095 0.8514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5095 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5095 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7950 2.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7950 2.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0805 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0805 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1496 2.0460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1496 2.0460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1625 -0.9176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1625 -0.9176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1339 0.7379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1339 0.7379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1606 -1.5971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1606 -1.5971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7950 2.8201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7950 2.8201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1744 0.4674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1744 0.4674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1886 -2.3038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1886 -2.3038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9209 -2.6843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9209 -2.6843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4814 -2.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4814 -2.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2724 -1.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2724 -1.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5401 -1.4624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5401 -1.4624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9795 -2.0681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9795 -2.0681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4997 -1.6291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4997 -1.6291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9460 -1.6536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9460 -1.6536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8641 -2.8201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8641 -2.8201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3475 -2.6628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3475 -2.6628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9652 -2.5212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9652 -2.5212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2690 -1.5815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2690 -1.5815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8563 -2.2962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8563 -2.2962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0580 -2.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0580 -2.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7355 -2.3139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7355 -2.3139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3229 -1.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3229 -1.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4755 -1.8082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4755 -1.8082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6324 -1.2224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6324 -1.2224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7522 -2.0647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7522 -2.0647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3050 -2.0371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3050 -2.0371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2295 -2.7270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2295 -2.7270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2110 -0.9795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2110 -0.9795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6409 -0.2674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6409 -0.2674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2289 0.4461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2289 0.4461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4647 -0.2674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4647 -0.2674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8767 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8767 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7006 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7006 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1131 -0.2665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1131 -0.2665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9381 -0.2665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9381 -0.2665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3506 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3506 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9381 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9381 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1131 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1131 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1744 -0.9809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1744 -0.9809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 13 22 1 0 0 0 0 | + | 13 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 18 1 0 0 0 0 | + | 26 18 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 36 43 1 0 0 0 0 | + | 36 43 1 0 0 0 0 |
| − | 20 37 1 0 0 0 0 | + | 20 37 1 0 0 0 0 |
| − | 44 40 1 0 0 0 0 | + | 44 40 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 54 49 1 0 0 0 0 | + | 54 49 1 0 0 0 0 |
| − | 52 55 1 0 0 0 0 | + | 52 55 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAGGL0069 | + | ID FL7AAGGL0069 |
| − | KNApSAcK_ID C00014820 | + | KNApSAcK_ID C00014820 |
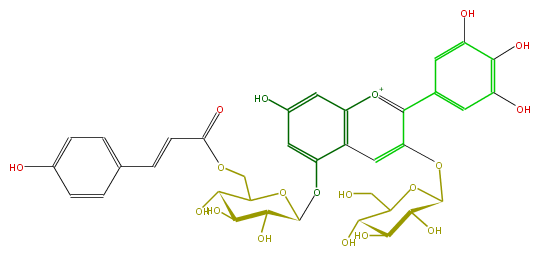
| − | NAME Delphinidin 3-glucoside-5-(6-(E)-p-coumaroylglucoside) | + | NAME Delphinidin 3-glucoside-5-(6-(E)-p-coumaroylglucoside) |
| − | CAS_RN 178737-74-1 | + | CAS_RN 178737-74-1 |
| − | FORMULA C36H37O19 | + | FORMULA C36H37O19 |
| − | EXACTMASS 773.192904002 | + | EXACTMASS 773.192904002 |
| − | AVERAGEMASS 773.66758 | + | AVERAGEMASS 773.66758 |
| − | SMILES OC(C6CO)C(C(O)C(O6)Oc(c5)c([o+1]c(c25)cc(cc2OC(C4O)OC(C(O)C4O)COC(C=Cc(c3)ccc(c3)O)=O)O)c(c1)cc(O)c(c1O)O)O | + | SMILES OC(C6CO)C(C(O)C(O6)Oc(c5)c([o+1]c(c25)cc(cc2OC(C4O)OC(C(O)C4O)COC(C=Cc(c3)ccc(c3)O)=O)O)c(c1)cc(O)c(c1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
0.4461 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4461 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1606 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8750 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8750 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1606 0.8155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5895 -0.8345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3040 -0.4220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3040 0.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5895 0.8155 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
4.0805 0.8514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7950 0.4389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5095 0.8514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5095 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7950 2.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0805 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1496 2.0460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1625 -0.9176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1339 0.7379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1606 -1.5971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7950 2.8201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1744 0.4674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1886 -2.3038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9209 -2.6843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4814 -2.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2724 -1.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5401 -1.4624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9795 -2.0681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4997 -1.6291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9460 -1.6536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8641 -2.8201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3475 -2.6628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9652 -2.5212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2690 -1.5815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8563 -2.2962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0580 -2.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7355 -2.3139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3229 -1.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4755 -1.8082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6324 -1.2224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7522 -2.0647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3050 -2.0371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2295 -2.7270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2110 -0.9795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6409 -0.2674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2289 0.4461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4647 -0.2674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8767 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7006 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1131 -0.2665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9381 -0.2665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3506 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9381 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1131 -1.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1744 -0.9809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
8 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
15 21 1 0 0 0 0
13 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 18 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
36 43 1 0 0 0 0
20 37 1 0 0 0 0
44 40 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
54 49 1 0 0 0 0
52 55 1 0 0 0 0
44 45 1 0 0 0 0
S SKP 8
ID FL7AAGGL0069
KNApSAcK_ID C00014820
NAME Delphinidin 3-glucoside-5-(6-(E)-p-coumaroylglucoside)
CAS_RN 178737-74-1
FORMULA C36H37O19
EXACTMASS 773.192904002
AVERAGEMASS 773.66758
SMILES OC(C6CO)C(C(O)C(O6)Oc(c5)c([o+1]c(c25)cc(cc2OC(C4O)OC(C(O)C4O)COC(C=Cc(c3)ccc(c3)O)=O)O)c(c1)cc(O)c(c1O)O)O
M END