Mol:FL7ARXGL0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 51 0 0 0 0 0 0 0 0999 V2000 | + | 46 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3533 -1.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -1.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6496 -0.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6496 -0.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6496 0.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6496 0.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0569 0.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0569 0.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0569 -0.7302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0569 -0.7302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0569 1.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0569 1.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7606 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7606 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7606 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7606 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0541 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0541 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0541 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0541 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6496 1.7074 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.6496 1.7074 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8374 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8374 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 1.3549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 1.3549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2176 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2176 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2176 2.5502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2176 2.5502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 2.9486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 2.9486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8374 2.5502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8374 2.5502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3337 1.6320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3337 1.6320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 -1.7808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -1.7808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7207 0.1037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7207 0.1037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 3.4835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 3.4835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9855 3.9415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9855 3.9415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6259 2.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6259 2.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6733 1.4902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6733 1.4902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3337 1.4902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3337 1.4902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7366 -0.4175 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.7366 -0.4175 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9191 -0.5281 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9191 -0.5281 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.5836 -1.0170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5836 -1.0170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9062 -1.7763 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9062 -1.7763 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7238 -1.6658 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7238 -1.6658 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0593 -1.1768 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.0593 -1.1768 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0300 -2.7686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0300 -2.7686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9513 -3.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9513 -3.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5697 -2.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5697 -2.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5778 -1.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5778 -1.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1832 -0.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1832 -0.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0441 -2.1797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0441 -2.1797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0441 -2.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0441 -2.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 -3.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -3.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6624 -2.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6624 -2.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6624 -2.1797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6624 -2.1797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 -3.9415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -3.9415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2451 -3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2451 -3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4650 -3.3948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4650 -3.3948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 3 11 2 0 0 0 0 | + | 3 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 7 1 0 0 0 0 | + | 13 7 1 0 0 0 0 |
| − | 12 14 1 0 0 0 0 | + | 12 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 9 20 1 0 0 0 0 | + | 9 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 11 22 1 0 0 0 0 | + | 11 22 1 0 0 0 0 |
| − | 18 23 1 0 0 0 0 | + | 18 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 17 25 1 0 0 0 0 | + | 17 25 1 0 0 0 0 |
| − | 16 26 1 0 0 0 0 | + | 16 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 31 34 1 0 0 0 0 | + | 31 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 28 38 1 0 0 0 0 | + | 28 38 1 0 0 0 0 |
| − | 29 22 1 0 0 0 0 | + | 29 22 1 0 0 0 0 |
| − | 21 39 2 0 0 0 0 | + | 21 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 43 21 1 0 0 0 0 | + | 43 21 1 0 0 0 0 |
| − | 41 44 1 0 0 0 0 | + | 41 44 1 0 0 0 0 |
| − | 42 45 1 0 0 0 0 | + | 42 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 34 35 | + | M SAL 4 2 34 35 |
| − | M SBL 4 1 37 | + | M SBL 4 1 37 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 37 2.5197 -2.0279 | + | M SVB 4 37 2.5197 -2.0279 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 45 46 | + | M SAL 3 2 45 46 |
| − | M SBL 3 1 50 | + | M SBL 3 1 50 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 50 -0.2451 -3.3948 | + | M SVB 3 50 -0.2451 -3.3948 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 26 27 | + | M SAL 2 2 26 27 |
| − | M SBL 2 1 29 | + | M SBL 2 1 29 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 29 2.6733 1.4902 | + | M SVB 2 29 2.6733 1.4902 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 23 24 | + | M SAL 1 2 23 24 |
| − | M SBL 1 1 26 | + | M SBL 1 1 26 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 26 1.5275 3.4835 | + | M SVB 1 26 1.5275 3.4835 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7ARXGL0006 | + | ID FL7ARXGL0006 |
| − | KNApSAcK_ID C00011189 | + | KNApSAcK_ID C00011189 |
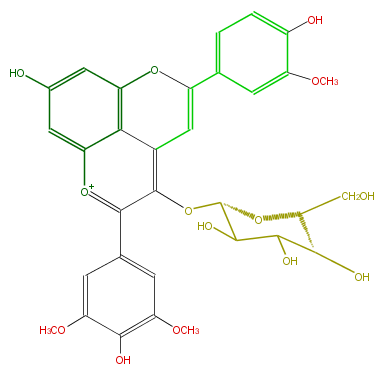
| − | NAME Malvidin 3-glucoside-4-vinylguaiacol | + | NAME Malvidin 3-glucoside-4-vinylguaiacol |
| − | CAS_RN 388089-40-5 | + | CAS_RN 388089-40-5 |
| − | FORMULA C32H31O14 | + | FORMULA C32H31O14 |
| − | EXACTMASS 639.1713807 | + | EXACTMASS 639.1713807 |
| − | AVERAGEMASS 639.58014 | + | AVERAGEMASS 639.58014 |
| − | SMILES O(c(c(c(c6)cc(OC)c(c(OC)6)O)5)c(C=2)c(c4[o+1]5)c(cc(c4)O)OC2c(c3)cc(OC)c(c3)O)[C@H](O1)C(O)C(O)[C@@H](O)[C@H]1CO | + | SMILES O(c(c(c(c6)cc(OC)c(c(OC)6)O)5)c(C=2)c(c4[o+1]5)c(cc(c4)O)OC2c(c3)cc(OC)c(c3)O)[C@H](O1)C(O)C(O)[C@@H](O)[C@H]1CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 51 0 0 0 0 0 0 0 0999 V2000
-1.3533 -1.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6496 -0.7302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6496 0.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0569 0.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0569 -0.7302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0569 1.7074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7606 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7606 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0541 0.4886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0541 1.3011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6496 1.7074 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.8374 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 1.3549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2176 1.7533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2176 2.5502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 2.9486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8374 2.5502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3337 1.6320 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 -1.7808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7207 0.1037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 3.4835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9855 3.9415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6259 2.7859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6733 1.4902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3337 1.4902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7366 -0.4175 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9191 -0.5281 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5836 -1.0170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9062 -1.7763 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7238 -1.6658 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0593 -1.1768 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0300 -2.7686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9513 -3.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5697 -2.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5778 -1.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1832 -0.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0441 -2.1797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0441 -2.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 -3.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6624 -2.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6624 -2.1797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 -3.9415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2451 -3.3948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4650 -3.3948 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
3 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 7 1 0 0 0 0
12 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
9 20 1 0 0 0 0
1 21 1 0 0 0 0
11 22 1 0 0 0 0
18 23 1 0 0 0 0
23 24 1 0 0 0 0
17 25 1 0 0 0 0
16 26 1 0 0 0 0
26 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
31 30 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
31 34 1 0 0 0 0
34 35 1 0 0 0 0
32 36 1 0 0 0 0
33 37 1 0 0 0 0
28 38 1 0 0 0 0
29 22 1 0 0 0 0
21 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
43 21 1 0 0 0 0
41 44 1 0 0 0 0
42 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 34 35
M SBL 4 1 37
M SMT 4 CH2OH
M SVB 4 37 2.5197 -2.0279
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 45 46
M SBL 3 1 50
M SMT 3 OCH3
M SVB 3 50 -0.2451 -3.3948
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 26 27
M SBL 2 1 29
M SMT 2 OCH3
M SVB 2 29 2.6733 1.4902
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 23 24
M SBL 1 1 26
M SMT 1 OCH3
M SVB 1 26 1.5275 3.4835
S SKP 8
ID FL7ARXGL0006
KNApSAcK_ID C00011189
NAME Malvidin 3-glucoside-4-vinylguaiacol
CAS_RN 388089-40-5
FORMULA C32H31O14
EXACTMASS 639.1713807
AVERAGEMASS 639.58014
SMILES O(c(c(c(c6)cc(OC)c(c(OC)6)O)5)c(C=2)c(c4[o+1]5)c(cc(c4)O)OC2c(c3)cc(OC)c(c3)O)[C@H](O1)C(O)C(O)[C@@H](O)[C@H]1CO
M END