Mol:FL7DAAGO0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4793 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4793 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4793 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4793 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0770 -0.1193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0770 -0.1193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6333 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6333 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6333 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6333 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0770 1.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0770 1.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1896 -0.1193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1896 -0.1193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7459 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7459 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7459 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7459 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1896 1.1654 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 1.1896 1.1654 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3020 1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3020 1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8690 0.8380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8690 0.8380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4359 1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4359 1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4359 1.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4359 1.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8690 2.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8690 2.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3020 1.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3020 1.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0027 2.1472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0027 2.1472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0354 1.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0354 1.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0770 -0.7614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0770 -0.7614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4262 0.9181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4262 0.9181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9106 0.2375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9106 0.2375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1681 0.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1681 0.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4517 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4517 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9723 1.0547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9723 1.0547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7307 0.7824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7307 0.7824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0027 0.5853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0027 0.5853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7144 0.1984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7144 0.1984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7427 -0.1880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7427 -0.1880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5187 -1.0413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5187 -1.0413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0031 -1.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0031 -1.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2606 -1.4331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2606 -1.4331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5442 -1.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5442 -1.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0648 -0.9047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0648 -0.9047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8232 -1.1770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8232 -1.1770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0952 -1.3741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0952 -1.3741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8069 -1.7610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8069 -1.7610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8352 -2.1473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8352 -2.1473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1188 -0.3696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1188 -0.3696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4043 -0.7821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4043 -0.7821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0263 1.5898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0263 1.5898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3118 1.1773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3118 1.1773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 19 1 0 0 0 0 | + | 32 19 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 42 -4.8339 5.5498 | + | M SBV 1 42 -4.8339 5.5498 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 44 -4.8339 5.5498 | + | M SBV 2 44 -4.8339 5.5498 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7DAAGS0003 | + | ID FL7DAAGS0003 |
| − | KNApSAcK_ID C00006622 | + | KNApSAcK_ID C00006622 |
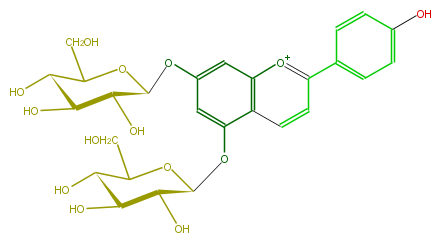
| − | NAME Apigeninidin 5,7-diglucoside | + | NAME Apigeninidin 5,7-diglucoside |
| − | CAS_RN 53948-04-2 | + | CAS_RN 53948-04-2 |
| − | FORMULA C27H31O14 | + | FORMULA C27H31O14 |
| − | EXACTMASS 579.1713807 | + | EXACTMASS 579.1713807 |
| − | AVERAGEMASS 579.52664 | + | AVERAGEMASS 579.52664 |
| − | SMILES c(c3)(OC(O5)C(C(C(O)C5CO)O)O)c(c(cc3OC(C4O)OC(CO)C(C(O)4)O)1)ccc(c(c2)ccc(O)c2)[o+1]1 | + | SMILES c(c3)(OC(O5)C(C(C(O)C5CO)O)O)c(c(cc3OC(C4O)OC(CO)C(C(O)4)O)1)ccc(c(c2)ccc(O)c2)[o+1]1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.4793 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4793 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0770 -0.1193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6333 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6333 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0770 1.1654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1896 -0.1193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7459 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7459 0.8442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1896 1.1654 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
2.3020 1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8690 0.8380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4359 1.1653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4359 1.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8690 2.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3020 1.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0027 2.1472 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0354 1.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0770 -0.7614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4262 0.9181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9106 0.2375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1681 0.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4517 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9723 1.0547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7307 0.7824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0027 0.5853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7144 0.1984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7427 -0.1880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5187 -1.0413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0031 -1.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2606 -1.4331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5442 -1.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0648 -0.9047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8232 -1.1770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0952 -1.3741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8069 -1.7610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8352 -2.1473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1188 -0.3696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4043 -0.7821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0263 1.5898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3118 1.1773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 18 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 19 1 0 0 0 0
34 38 1 0 0 0 0
38 39 1 0 0 0 0
25 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 42
M SMT 1 ^CH2OH
M SBV 1 42 -4.8339 5.5498
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 ^CH2OH
M SBV 2 44 -4.8339 5.5498
S SKP 8
ID FL7DAAGS0003
KNApSAcK_ID C00006622
NAME Apigeninidin 5,7-diglucoside
CAS_RN 53948-04-2
FORMULA C27H31O14
EXACTMASS 579.1713807
AVERAGEMASS 579.52664
SMILES c(c3)(OC(O5)C(C(C(O)C5CO)O)O)c(c(cc3OC(C4O)OC(CO)C(C(O)4)O)1)ccc(c(c2)ccc(O)c2)[o+1]1
M END