Mol:FLIA1ADS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2970 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2970 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2970 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2970 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7407 -1.0642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7407 -1.0642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1844 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1844 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1844 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1844 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7407 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7407 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6281 -1.0642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6281 -1.0642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0718 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0718 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0718 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0718 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6281 0.2205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6281 0.2205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6281 -1.5650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6281 -1.5650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8531 0.2204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8531 0.2204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5159 2.6987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5159 2.6987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1215 2.2400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1215 2.2400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8646 1.5793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8646 1.5793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8577 0.9419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8577 0.9419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3944 1.4051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3944 1.4051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6994 1.9830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6994 1.9830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6777 3.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6777 3.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1563 2.9552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1563 2.9552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5000 1.2008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5000 1.2008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4538 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4538 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1324 -2.0774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1324 -2.0774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7186 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7186 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7186 -1.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7186 -1.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1324 -0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1324 -0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4538 -1.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4538 -1.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1442 -2.1646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1442 -2.1646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2965 -2.2663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2965 -2.2663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7603 -1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7603 -1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1521 -2.6788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1521 -2.6788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7296 -2.4107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7296 -2.4107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3278 -2.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3278 -2.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8534 -1.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8534 -1.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9619 -1.6867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9619 -1.6867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5733 -2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5733 -2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8531 -2.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8531 -2.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8140 -3.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8140 -3.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0171 -3.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0171 -3.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1841 2.2806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1841 2.2806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4696 1.8681 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4696 1.8681 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 28 24 1 0 0 0 0 | + | 28 24 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 18 40 1 0 0 0 0 | + | 18 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 42 -8.2616 5.6074 | + | M SBV 1 42 -8.2616 5.6074 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 44 -7.8307 6.7969 | + | M SBV 2 44 -7.8307 6.7969 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIA1ADS0001 | + | ID FLIA1ADS0001 |
| − | KNApSAcK_ID C00006228 | + | KNApSAcK_ID C00006228 |
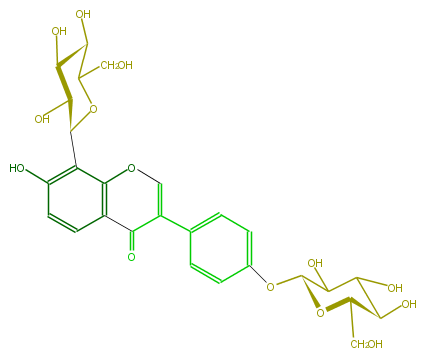
| − | NAME Puerarin 4'-O-glucoside | + | NAME Puerarin 4'-O-glucoside |
| − | CAS_RN 117047-08-2 | + | CAS_RN 117047-08-2 |
| − | FORMULA C27H30O14 | + | FORMULA C27H30O14 |
| − | EXACTMASS 578.163555668 | + | EXACTMASS 578.163555668 |
| − | AVERAGEMASS 578.5187000000001 | + | AVERAGEMASS 578.5187000000001 |
| − | SMILES C(C5O)(CO)OC(C(O)C5O)c(c41)c(O)ccc1C(C(=CO4)c(c3)ccc(c3)OC(O2)C(O)C(O)C(O)C2CO)=O | + | SMILES C(C5O)(CO)OC(C(O)C5O)c(c41)c(O)ccc1C(C(=CO4)c(c3)ccc(c3)OC(O2)C(O)C(O)C(O)C2CO)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-3.2970 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2970 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7407 -1.0642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1844 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1844 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7407 0.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6281 -1.0642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0718 -0.7430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0718 -0.1007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6281 0.2205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6281 -1.5650 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8531 0.2204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5159 2.6987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1215 2.2400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8646 1.5793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8577 0.9419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3944 1.4051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6994 1.9830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6777 3.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1563 2.9552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5000 1.2008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4538 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1324 -2.0774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7186 -1.7390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7186 -1.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1324 -0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4538 -1.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1442 -2.1646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2965 -2.2663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7603 -1.9569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1521 -2.6788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7296 -2.4107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3278 -2.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8534 -1.9775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9619 -1.6867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5733 -2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8531 -2.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8140 -3.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0171 -3.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1841 2.2806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4696 1.8681 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 22 1 0 0 0 0
27 8 1 0 0 0 0
28 24 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
30 28 1 0 0 0 0
32 38 1 0 0 0 0
38 39 1 0 0 0 0
18 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 42
M SMT 1 CH2OH
M SBV 1 42 -8.2616 5.6074
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 CH2OH
M SBV 2 44 -7.8307 6.7969
S SKP 8
ID FLIA1ADS0001
KNApSAcK_ID C00006228
NAME Puerarin 4'-O-glucoside
CAS_RN 117047-08-2
FORMULA C27H30O14
EXACTMASS 578.163555668
AVERAGEMASS 578.5187000000001
SMILES C(C5O)(CO)OC(C(O)C5O)c(c41)c(O)ccc1C(C(=CO4)c(c3)ccc(c3)OC(O2)C(O)C(O)C(O)C2CO)=O
M END