Mol:FLIAABGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7298 1.4107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7298 1.4107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0447 1.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0447 1.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6833 1.5995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6833 1.5995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4108 1.1795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4108 1.1795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4108 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4108 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1657 -0.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1657 -0.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9203 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9203 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9203 1.1795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9203 1.1795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1657 1.6151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1657 1.6151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0445 0.3391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0445 0.3391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6833 -0.0811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6833 -0.0811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1707 -0.9606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1707 -0.9606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6747 -0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6747 -0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6747 -0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6747 -0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3960 -1.3772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3960 -1.3772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1177 -0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1177 -0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1177 -0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1177 -0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3960 0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3960 0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3172 1.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3172 1.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8138 0.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8138 0.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0888 1.1345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0888 1.1345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3332 1.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3332 1.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8976 1.6506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8976 1.6506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6381 1.3845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6381 1.3845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8595 1.2768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8595 1.2768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4350 0.8593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4350 0.8593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5099 0.7135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5099 0.7135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1405 1.8245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1405 1.8245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4655 1.9291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4655 1.9291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9621 1.2644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9621 1.2644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2370 1.5463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2370 1.5463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4815 1.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4815 1.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0460 2.0624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0460 2.0624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7864 1.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7864 1.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.8856 1.8338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.8856 1.8338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6447 1.2887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6447 1.2887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6118 1.1022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6118 1.1022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8156 1.8229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8156 1.8229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6696 -0.9195 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6696 -0.9195 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7524 -1.4080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7524 -1.4080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.8856 -2.0624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.8856 -2.0624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 38 28 1 0 0 0 0 | + | 38 28 1 0 0 0 0 |
| − | 11 39 1 0 0 0 0 | + | 11 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 16 40 1 0 0 0 0 | + | 16 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 45 -0.6347 0.4473 | + | M SBV 1 45 -0.6347 0.4473 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FLIAABGS0004 | + | ID FLIAABGS0004 |
| − | FORMULA C27H30O14 | + | FORMULA C27H30O14 |
| − | EXACTMASS 578.163555668 | + | EXACTMASS 578.163555668 |
| − | AVERAGEMASS 578.5187000000001 | + | AVERAGEMASS 578.5187000000001 |
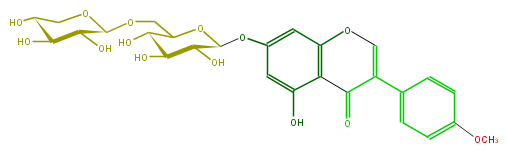
| − | SMILES OC(C(COC(O5)C(O)C(C(O)C5)O)4)C(C(C(O4)Oc(c3)cc(c(c32)C(=O)C(=CO2)c(c1)ccc(OC)c1)O)O)O | + | SMILES OC(C(COC(O5)C(O)C(C(O)C5)O)4)C(C(C(O4)Oc(c3)cc(c(c32)C(=O)C(=CO2)c(c1)ccc(OC)c1)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.7298 1.4107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0447 1.1793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6833 1.5995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4108 1.1795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4108 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1657 -0.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9203 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9203 1.1795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1657 1.6151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0445 0.3391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6833 -0.0811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1707 -0.9606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6747 -0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6747 -0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3960 -1.3772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1177 -0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1177 -0.1276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3960 0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3172 1.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8138 0.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0888 1.1345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3332 1.1262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8976 1.6506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6381 1.3845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8595 1.2768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4350 0.8593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5099 0.7135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1405 1.8245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.4655 1.9291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9621 1.2644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2370 1.5463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4815 1.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0460 2.0624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.7864 1.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.8856 1.8338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.6447 1.2887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6118 1.1022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8156 1.8229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6696 -0.9195 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7524 -1.4080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.8856 -2.0624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
24 28 1 0 0 0 0
22 1 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
38 28 1 0 0 0 0
11 39 1 0 0 0 0
40 41 1 0 0 0 0
16 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 OCH3
M SBV 1 45 -0.6347 0.4473
S SKP 5
ID FLIAABGS0004
FORMULA C27H30O14
EXACTMASS 578.163555668
AVERAGEMASS 578.5187000000001
SMILES OC(C(COC(O5)C(O)C(C(O)C5)O)4)C(C(C(O4)Oc(c3)cc(c(c32)C(=O)C(=CO2)c(c1)ccc(OC)c1)O)O)O
M END